28S rDNA C-Region:The 28S C-DNA is universally suitable for all sponge classes (including calcareous sponges) and provides a higher resolution and easier amplification [1,2] with the current barcoding primers designed by Chombard et al. 1998 [3]:
Forward: C2: 5′-GAAAAGAACTTTGRARAGAGAGT-3′
Reverse: D2: 5′-TCCGTGTTTCAAGACGGG-3′
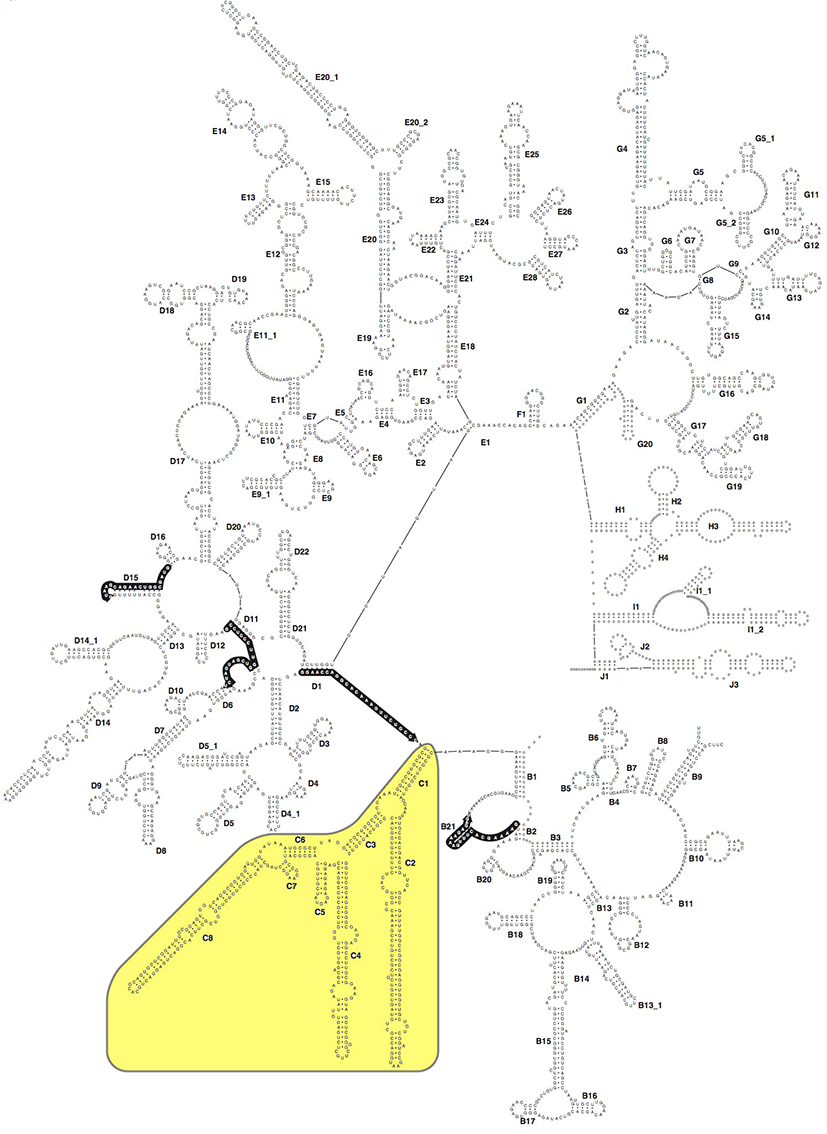 Fig. 1: 28S rDNA secondary structure, the C-Region is highlighted in yellow. Modified after [1].
Fig. 1: 28S rDNA secondary structure, the C-Region is highlighted in yellow. Modified after [1].
CO1 standard barcoding fragmentThe standard barcoding fragment for the Barcoding of Life initiative is a approx. 640 bp long fragment located at the 5' site of the mitochondrial cytochrome oxidase subunit 1 (CO1). CO1 in Calcarea, however canot be amplified with standardized primers due to their high mitochondrial substitution rates [4]. For all other sponge classes CO1 is best amplified with the degenerated (standard) CO1 barcoding primer designed by Meyer et al. [5]:
Forward: dgLCO1490: 5′-GGT CAA CAA ATC ATA AAG AYA TYG G
Reverse: dgHCO2198: 5′-TAA ACT TCA GGG TGA CCA AAR AAY CA
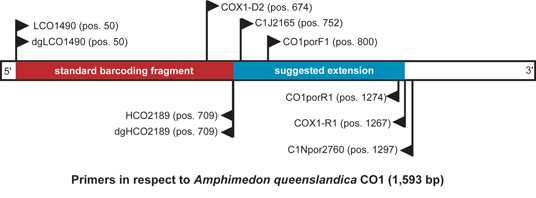 Fig. 2: Schematic view of the CO1 standard barcoding region (Folmer et al. 1994 [6]), and the extension region ("I3M11", Erpenbeck et al. [7]) with the mentioned primers .
Fig. 2: Schematic view of the CO1 standard barcoding region (Folmer et al. 1994 [6]), and the extension region ("I3M11", Erpenbeck et al. [7]) with the mentioned primers .
CO1 extension region (I3M11)The standard barcoding fragment might not display sufficient variability in sponges to species level [7]. An additional downstream region (I3M11) exhibits a higher substitution rate suitable for population studies. The following primers for the extended fragment are designed by Misof et al. [8] and Erpenbeck et al. [7]:
Forward: CO1porF1: 5′-CCN CAN TTN KCN GMN AAA AAA CA-3′ [7]
Reverse: CO1porR1: 5′-AAN TGN TGN GGR AAR AAN G-3′ [7]
Forward: C1J2165: 5′-GAA GTT TAT ATT TTA ATT TTA CCN GG-3′ [8]
Reverse: C1Npor2760: 5′-TCT AGG TAA TCC AGC TAA ACC-3′ [8]
NOTE: We suggest a nested approach in which a C1J2165/C1Npor2760 PCR-product will be reamplified by CO1porF1/CO1porR1. See [9] for details.
A further primer set is developed by Rot et al. [10], see Fig. 2.
Forward: COX1-D2: 5′-AAT ACT GCT TTT TTT GAT CCT GCC GG-3′
Reverse: COX1-R1: 5′-TGT TGR GGG AAA AAR GTT AAA TT-3′
Some species (including many Tetillidae) possess an intron in the priming site for the standard reverse primer (dgHCO2198). The combination of dgLCO1490 (forward) with COX1-R1 (reverse) can help to bridge this intron.
References:
[1] Voigt O, Wörheide G (2016) A short LSU rRNA fragment as a standard marker for integrative taxonomy in calcareous sponges (Porifera: Calcarea). Org Divers Evol 16:53–64.
[2] Erpenbeck D, Voigt O, Al-Aidaroos AM, et al (2016) Molecular biodiversity of Red Sea demosponges. Mar Pollut Bull 105:507–514.
[3] Chombard C, Boury-Esnault N, Tillier S (1998) Reassessment of homology of morphological characters in tetractinellid sponges based on molecular data. Syst Biol 47:351–366.
[4] Lavrov D, Pett W, Voigt O, et al (2013) Mitochondrial DNA of Clathrina clathrus (Calcarea, Calcinea): six linear chromosomes, fragmented rRNAs, tRNA editing, and a novel genetic code. Mol Biol Evol 30:865–880.
[5] Meyer CP, Geller JB & Paulay G (2005) Fine scale endemism on coral reefs: Archipelagic differentiation in turbinid gastropods. Evolution 59: 113-125.
[6] Folmer O, Black M, Hoeh W, Lutz R & Vrijenhoek R (1994) DNA primers for amplification of mitochondrial cytochrome C oxidase subunit I from diverse metazoan invertebrates. :. Mol. Mar. Biol. Biotechnol. 3: 294-299.
[7] Erpenbeck D, Hooper JNA, Wörheide G (2006) CO1 phylogenies in diploblasts and the “Barcoding of Life” - are we sequencing a suboptimal partition? Mol Ecol Notes 6:550–553.
[8] Misof B, Erpenbeck D & Sauer KP (2000) Mitochondrial gene fragments suggest paraphyly of the genus Panorpa (Mecoptera, Panorpidae). Molecular Phylogenetics and Evolution 17: 76-84.
[9] Erpenbeck D, Knowlton AL, Talbot SL, Highsmith RC & Van Soest RWM (2003 (2004)) A molecular comparison of Alaskan and North East Atlantic Halichondria panicea (Pallas 1766) (Porifera: Demospongiae) populations. Boll. Mus. Ist. Univ. Genova. 68: 319-325.
[10] Rot C, Goldfarb I, Ilan M & Huchon D (2006) Putative cross-kingdom horizontal gene transfer in sponge (Porifera) mitochondria. BMC Evolutionary Biology 6.











